All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional. If you are a patient or carer, please visit the Lymphoma Coalition.
The lym Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the lym Hub cannot guarantee the accuracy of translated content. The lym and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The Lymphoma & CLL Hub is an independent medical education platform, sponsored by AbbVie, BeOne Medicines, Johnson & Johnson, Roche and sobi, and supported through educational grants from Bristol Myers Squibb, Incyte and Lilly. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View lymphoma & CLL content recommended for you
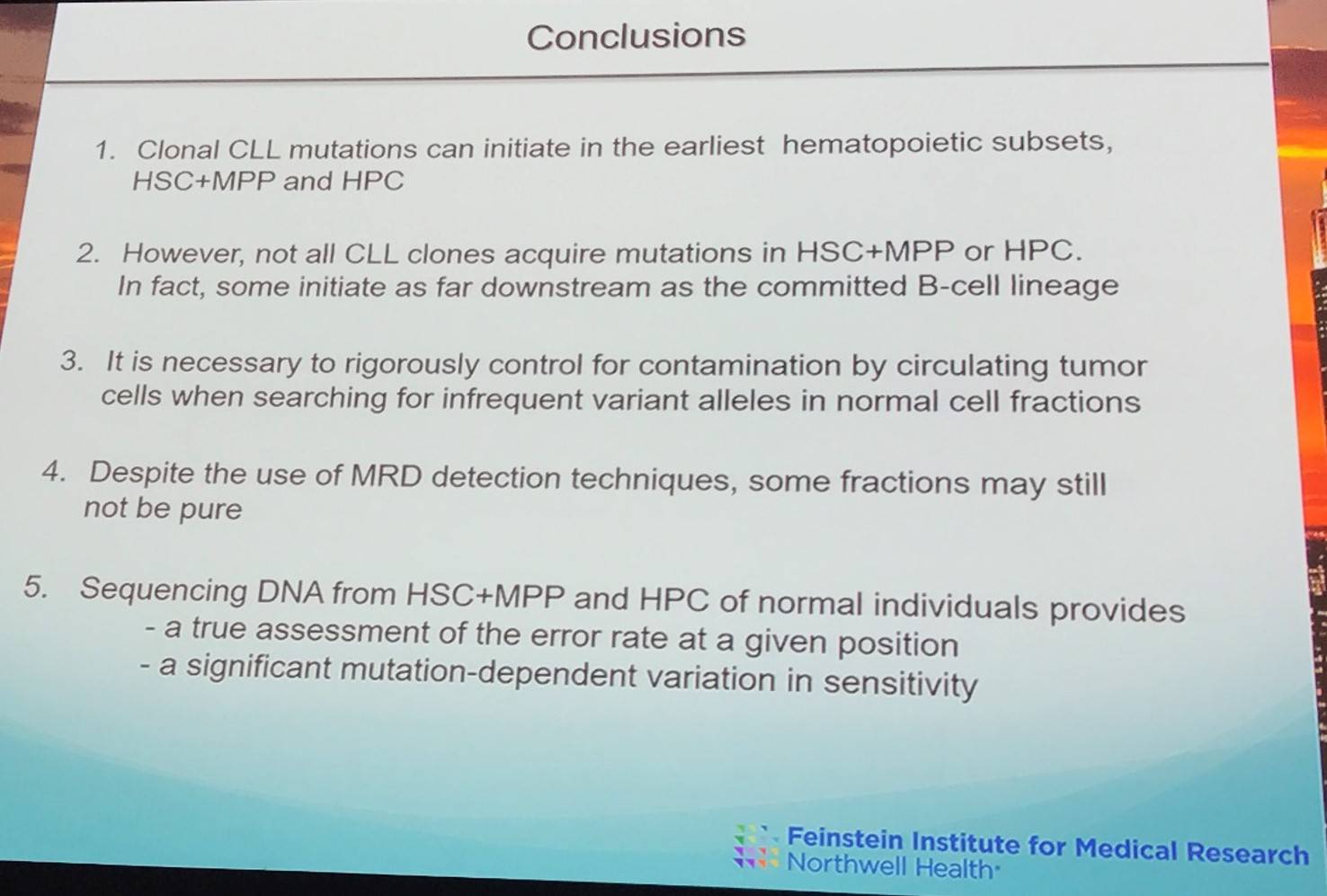
iwCLL 2017 | Clonal Chronic Lymphocytic Leukemia mutations can occur extremely early in the hematopoietic lineage
The second session at this year’s iwCLL was titled “Role of Non-Leukemic Cells and the Microenvironment in CLL Development”, and was jointly chaired by Silvia Deaglio (University of Turin, Torino, Italy) and Christopher Pepper (Cardiff University, Wales, UK).
Sonia Marsilio, PhD, from the Weill Cornell Medical College, New York, USA, gave a talk titled “CLL Mutations in the Earliest Cells of the Hematopoietic Lineage: Documentation and Need for Caution.”
The group aimed to analyze the genomes of immature members of the hematopoietic lineage, while preventing contamination from mature CLL cells and controlling for deep sequencing noise.
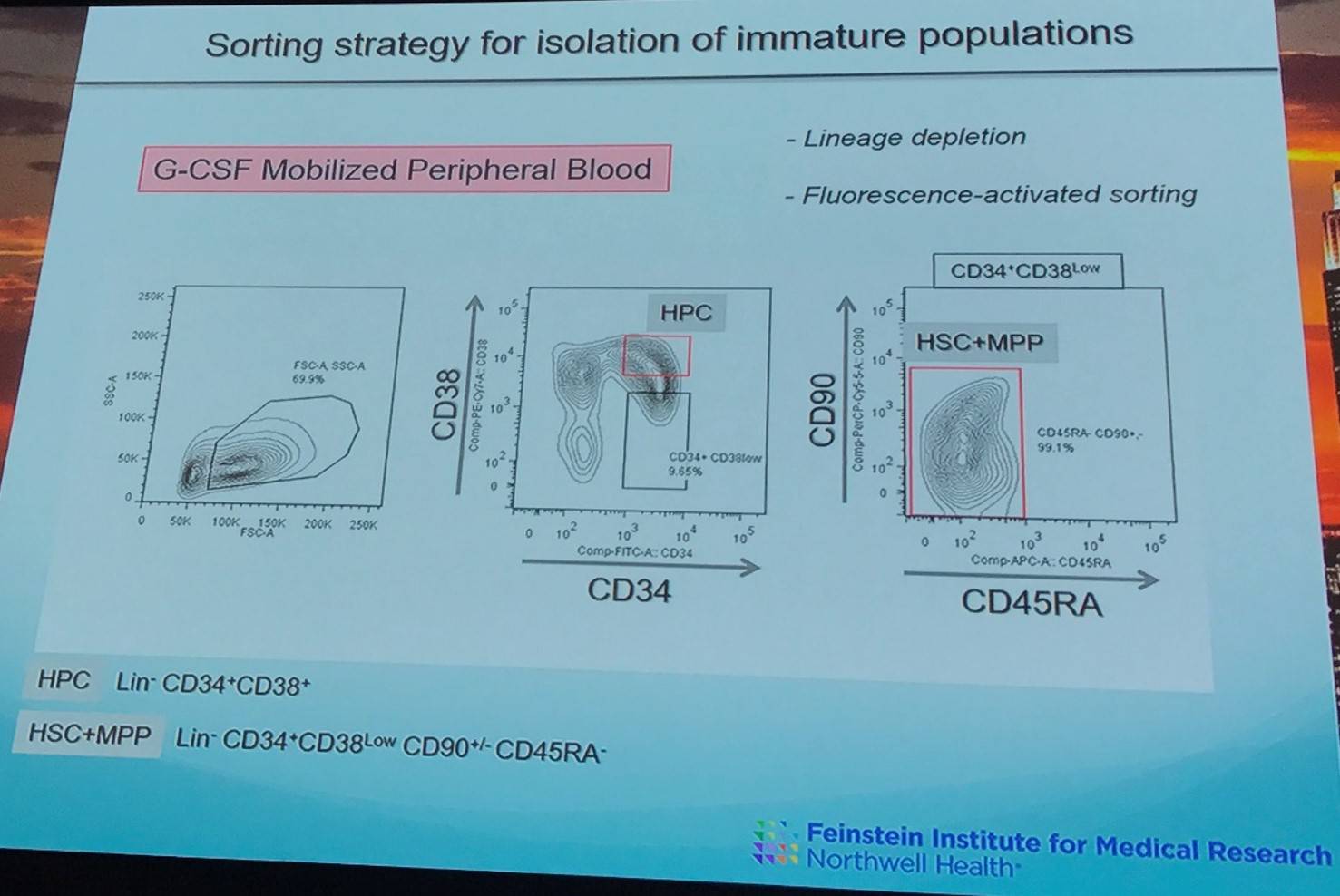
FACS was used to isolate cell populations containing Hematopoietic Stem Cells (HSCs), Multi-Potent Progenitor (MPP) cells, downstream Hematopoietic Progenitor Cell (HPC) populations, and mature T-cells and monocytes from granulocyte-colony stimulating factor mobilized peripheral blood of 9 patients. CLL cells were also identified and subjected to Whole Exome Sequencing (WES).
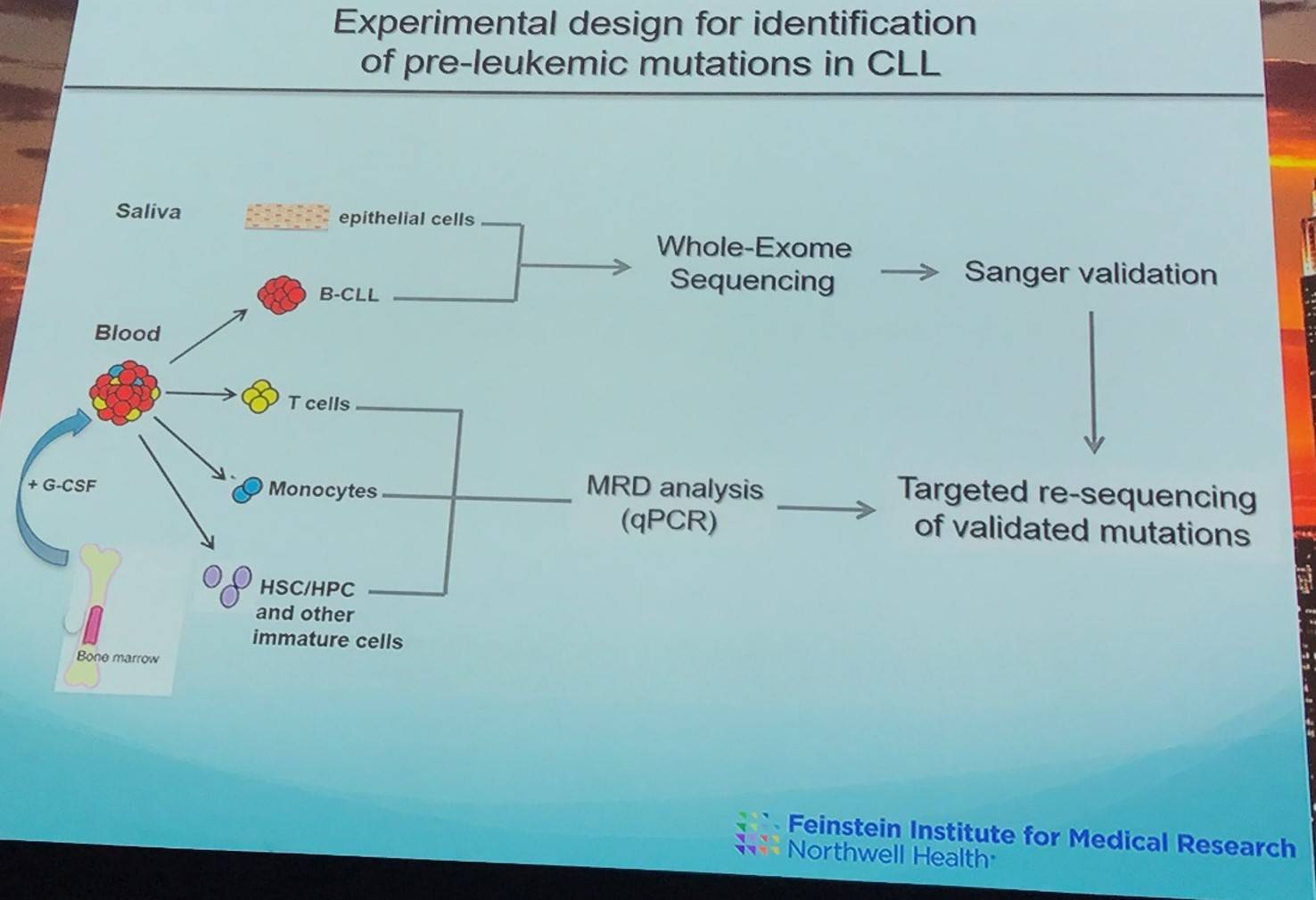
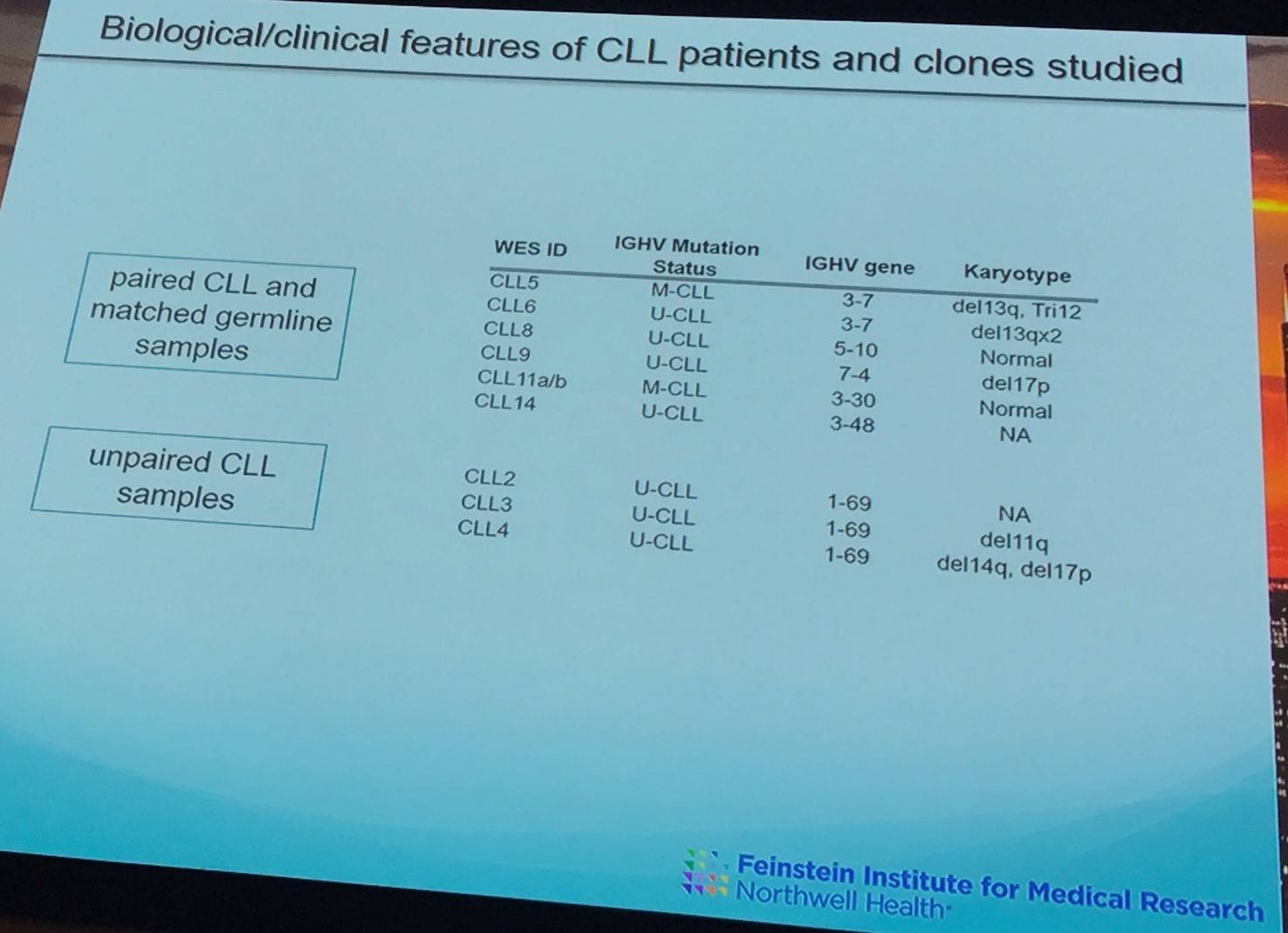
In 6 paired CLL samples, 87 coding mutation were identified (average = 14.5 mutations per sample) and went on to be validated by Sanger sequencing.
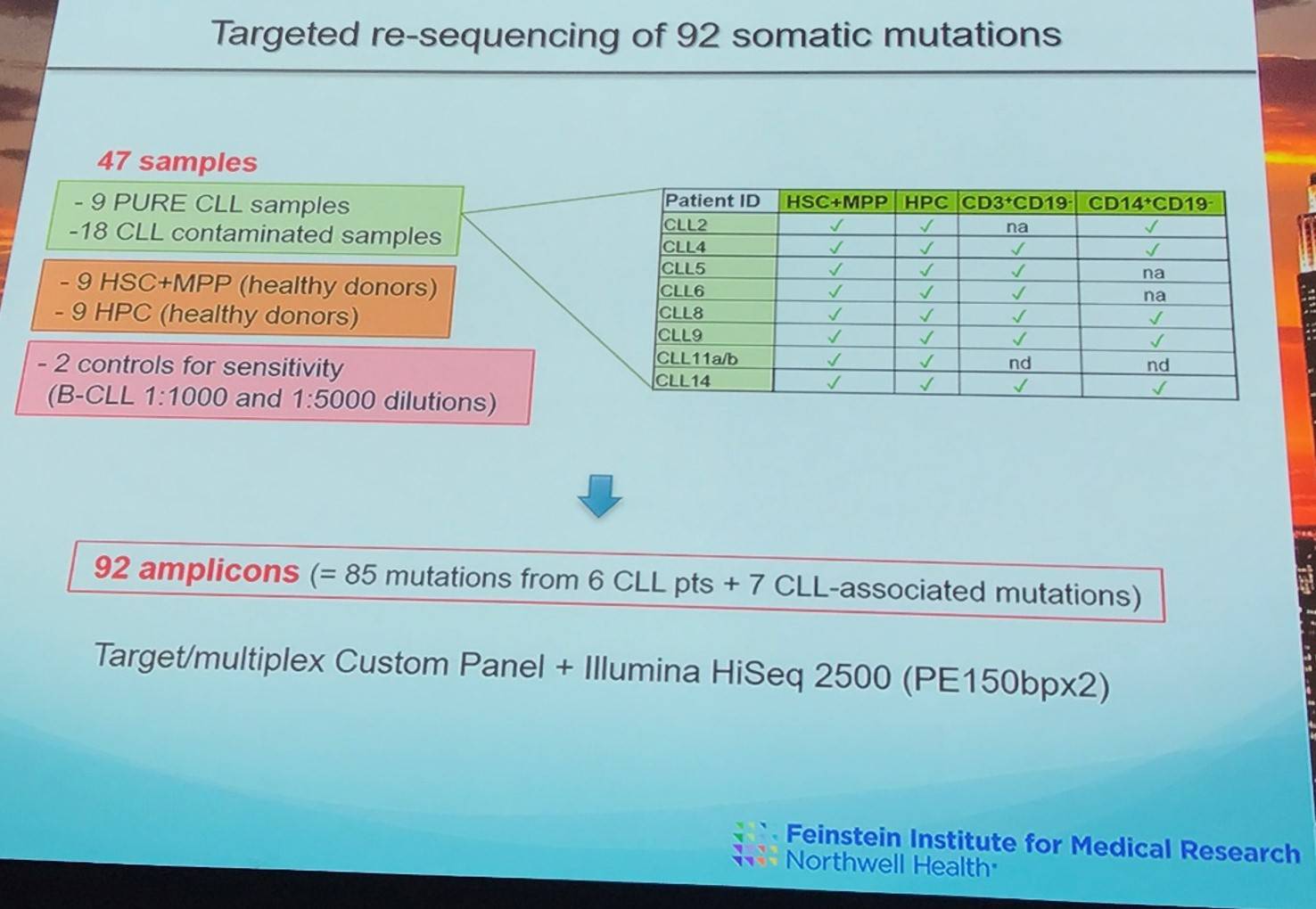
Using the MRD method defined by van der Velden et al. in Leukemia in 2007, 9 pure samples were identified (0.01–0.04% of B-CLL cells = 11; 0.05–1.4% B-CLL cells = 9).
In one patient, an SF3B1-K700E mutation was present in the HSC+MPP fraction (VAF >0.15%) as well as in CLL B-cells (VAF >0.15%) of the patient in mutually exclusive clones. The SF3B1-I704F mutation was only identified in later stages of hematopoietic differentiation (CLL B-cells, VAF = 47%).
However, in some cases HSC+MPP and HPC fractions defined as “pure” by MRD analysis potentially were still contaminated with CLL B-cells.
|
Pt ID |
HSC+MPP |
HPC |
T-Cells |
Monocytes |
Level of mutation |
|
|---|---|---|---|---|---|---|
|
CLL5 |
PURE |
- |
|
NA |
HSC+MPP |
50% of WES defined mutations |
|
CLL11 |
- |
PURE |
- |
- |
HPC |
93% of WES defined mutations |
Sonia Marsilio concluded her presentation with a succinct summary slide:
References
Your opinion matters
In your experience, when do most CRS/ICANS events occur after lisocabtagene maraleucel infusion?

